Adenosine diphosphate
 | |
 | |
| Names | |
|---|---|
IUPAC name [(2R,3S,4R,5R)-5-(6-Aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl phosphono hydrogen phosphate | |
| Other names Adenosine 5′-diphosphate; Adenosine 5′-pyrophosphate; Adenosine pyrophosphate | |
| Identifiers | |
CAS Number |
|
3D model (JSmol) |
|
ChEBI |
|
ChEMBL |
|
ChemSpider |
|
DrugBank |
|
ECHA InfoCard | 100.000.356 |
EC Number | 218-249-0 |
IUPHAR/BPS |
|
KEGG |
|
PubChem CID |
|
RTECS number | AU7467000 |
InChI
| |
SMILES
| |
| Properties | |
Chemical formula | C10H15N5O10P2 |
Molar mass | 427.201 g/mol |
| Appearance | white powder |
Density | 2.49 g/mL |
log P | -2.640 |
| Hazards | |
Safety data sheet | MSDS |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
Infobox references | |
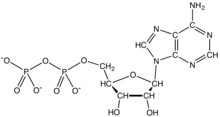
Adenosine diphosphate (ADP), also known as adenosine pyrophosphate (APP), is an important organic compound in metabolism and is essential to the flow of energy in living cells. ADP consists of three important structural components: a sugar backbone attached to adenine and two phosphate groups bonded to the 5 carbon atom of ribose. The diphosphate group of ADP is attached to the 5’ carbon of the sugar backbone, while the adenosine attaches to the 1’ carbon.[1]
ADP can be interconverted to adenosine triphosphate (ATP) and adenosine monophosphate (AMP). ATP contains one more phosphate group than does ADP. AMP contains one fewer phosphate group. Energy transfer used by all living things is a result of dephosphorylation of ATP by enzymes known as ATPases. The cleavage of a phosphate group from ATP results in the coupling of energy to metabolic reactions and a by-product of ADP.[1] Being the "molecular unit of currency", ATP is continually being reformed from lower-energy species ADP and AMP. The biosynthesis of ATP is achieved throughout processes such as substrate-level phosphorylation, oxidative phosphorylation, and photophosphorylation, all of which facilitating the addition of a phosphate group to ADP.
Contents
1 Bioenergetics
2 Cellular respiration
2.1 Catabolism
2.2 Glycolysis
2.3 Citric acid cycle
2.4 Oxidative phosphorylation
2.4.1 Mitochondrial ATP synthase complex
3 Blood platelet activation
4 See also
5 References
Bioenergetics
ADP cycling supplies the energy needed to do work in a biological system, the thermodynamic process of transferring energy from one source to another. There are two types of energy: potential energy and kinetic energy. Potential energy can be thought of as stored energy, or usable energy that is available to do work. Kinetic energy is the energy of an object as a result of its motion. The significance of ATP is in its ability to store potential energy within the phosphate bonds. The energy stored between these bonds can then be transferred to do work. For example, the transfer of energy from ATP to the protein myosin causes a conformational change when connecting to actin during muscle contraction. It takes multiple reactions between myosin and actin to effectively produce one muscle contraction, and, therefore, the availability of large amounts of ATP is required to produce each muscle contraction. For this reason, biological processes have evolved to produce efficient ways to replenishment the potential energy of ATP from ADP.[2]
Breaking one of ATP's phosphorus bonds generates approximately 30.5 kilojoules per Mole of ATP (7.3 kcal).[3] ADP can be converted, or powered back to ATP through the process of releasing the chemical energy available in food; in humans, this is constantly performed via aerobic respiration in the mitochondria.[2] Plants use photosynthetic pathways to convert and store energy from sunlight, also conversion of ADP to ATP.[3] Animals use the energy released in the breakdown of glucose and other molecules to convert ADP to ATP, which can then be used to fuel necessary growth and cell maintenance.[2]
Cellular respiration
Catabolism
The ten-step catabolic pathway of glycolysis is the initial phase of free-energy release in the breakdown of glucose and can be split into two phases, the preparatory phase and payoff phase. ADP and phosphate are needed as precursors to synthesize ATP in the payoff reactions of the TCA cycle and oxidative phosphorylation mechanism.[4] During the payoff phase of glycolysis, the enzymes phosphoglycerate kinase and pyruvate kinase facilitate the addition of a phosphate group to ADP by way of substrate-level phosphorylation.[5]

Glycolysis overview
Glycolysis
Glycolysis is performed by all living organisms and consists of 10 steps. The net reaction for the overall process of glycolysis is:[6]
- Glucose + 2 NAD+ + 2 Pi + 2 ADP → 2 pyruvate + 2 ATP + 2 NADH + 2 H2O
Steps 1 and 3 require the input of energy derived from the hydrolysis of ATP to ADP and Pi (inorganic phosphate), whereas steps 7 and 10 require the input of ADP, each yielding ATP.[7] The enzymes necessary to break down glucose are found in the cytoplasm, the viscous fluid that fills living cells, where the glycolytic reactions take place.
Citric acid cycle
The citric acid cycle, also known as the Krebs cycle or the TCA (tricarboxylic acid) cycle is an 8-step process that takes the pyruvate generated by glycolysis and generates 4 NADH, FADH2, and GTP, which is further converted to ATP.[8] It is only in step 5, where GTP is generated, by succinyl-CoA synthetase, and then converted to ATP, that ADP is used (GTP + ADP → GDP + ATP).[9]
Oxidative phosphorylation
Oxidative phosphorylation produces 26 of the 30 equivalents of ATP generated in cellular respiration by transferring electrons from NADH or FADH2 to O2 through electron carriers.[10] The energy released when electrons are passed from higher-energy NADH or FADH2 to the lower-energy O2 is required to phosphorylate ADP and once again generate ATP.[11] It is this energy coupling and phosphorylation of ADP to ATP that gives the electron transport chain the name oxidative phosphorylation.
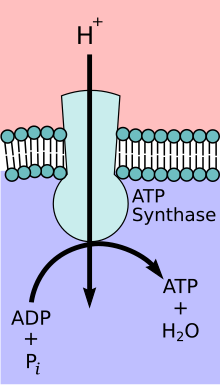
ATP-Synthase
Mitochondrial ATP synthase complex
During the initial phases of glycolysis and the TCA cycle, cofactors such as NAD+ donate and accept electrons[12] that aid in the electron transport chain's ability to produce a proton gradient across the inner mitochondrial membrane.[13] The ATP synthase complex exists within the mitochondrial membrane (F0 portion) and protrudes into the matrix (F1portion). The energy derived as a result of the chemical gradient is then used to synthesize ATP by coupling the reaction of inorganic phosphate to ADP in the active site of the ATP synthase enzyme; the equation for this can be written as ADP + Pi → ATP.
Blood platelet activation
Under normal conditions, small disk-shape platelets circulate in the blood freely and without interaction with one another. ADP is stored in dense bodies inside blood platelets and is released upon platelet activation. ADP interacts with a family of ADP receptors found on platelets (P2Y1, P2Y12, and P2X1), which leads to platelet activation.[14]
P2Y1 receptors initiate platelet aggregation and shape change as a result of interactions with ADP.
P2Y12 receptors further amplify the response to ADP and draw forth the completion of aggregation.
ADP in the blood is converted to adenosine by the action of ecto-ADPases, inhibiting further platelet activation via adenosine receptors.
See also
- Nucleoside
- Nucleotide
- DNA
- RNA
- Oligonucleotide
- Apyrase
- Phosphate
References
^ ab Cox, Michael; Nelson, David R.; Lehninger, Albert L (2008). Lehninger principles of biochemistry. San Francisco: W.H. Freeman. ISBN 0-7167-7108-X..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ abc Nave, C.R. (2005). "Adenosine Triphosphate". Hyper Physics [serial on the Internet]. Georgia State University.
^ ab Farabee, M.J. (2002). "The Nature of ATP". ATP and Biological Energy [serial on the Internet]. Archived from the original on 2007-12-01.
^ Jensen TE, Richter EA (March 2012). "Regulation of glucose and glycogen metabolism during and after exercise". J. Physiol. 590 (Pt 5): 1069–76. doi:10.1113/jphysiol.2011.224972. PMC 3381815. PMID 22199166.
^ Liapounova NA, Hampl V, Gordon PM, Sensen CW, Gedamu L, Dacks JB (December 2006). "Reconstructing the mosaic glycolytic pathway of the anaerobic eukaryote Monocercomonoides". Eukaryotic Cell. 5 (12): 2138–46. doi:10.1128/EC.00258-06. PMC 1694820. PMID 17071828.
^ Medh, J.D. "Glycolysis" (PDF). CSUN.Edu. Retrieved 3 April 2013.
^ Bailey, Regina. "10 Steps of Glycolysis".
^ "Citric Acid Cycle" (PDF). Takusagawa’s Note. Archived from the original (PDF) on 24 March 2012. Retrieved 4 April 2013.
^ "Biochemistry" (PDF). UCCS.edu. Archived from the original (PDF) on 2013-02-28.
^ "Oxidative phosphorylation". W H Freeman, 2002. Retrieved 4 April 2013.
^ Medh, J. D. "Electron Transport Chain (Overview)" (PDF). CSUN.edu. Retrieved 4 April 2013.
^ Belenky P, Bogan KL, Brenner C (January 2007). "NAD+ metabolism in health and disease". Trends Biochem. Sci. 32 (1): 12–9. doi:10.1016/j.tibs.2006.11.006. PMID 17161604.
^ Murray, Robert F. (2003). Harper's illustrated biochemistry. New York: McGraw-Hill. ISBN 0-07-121766-5.
^ Murugappa S, Kunapuli SP (2006). "The role of ADP receptors in platelet function". Front. Biosci. 11: 1977–86. doi:10.2741/1939. PMID 16368572.