Mycobacterium
Mycobacterium | |
|---|---|
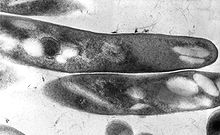 | |
TEM micrograph of M. tuberculosis. | |
Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Actinobacteria |
| Order: | Actinomycetales |
| Suborder: | Corynebacterineae |
| Family: | Mycobacteriaceae |
| Genus: | Mycobacterium Lehmann & Neumann 1896 |
Species | |
See below. | |
Mycobacterium is a genus of Actinobacteria, given its own family, the Mycobacteriaceae. Over 190 species are recognized in this genus.[1] This genus includes pathogens known to cause serious diseases in mammals, including tuberculosis (Mycobacterium tuberculosis) and leprosy (Mycobacterium leprae) in humans.[2] The Greek prefix myco- means "fungus," alluding to the way mycobacteria have been observed to grow in a mold-like fashion on the surface of cultures.[3]
It is acid fast and cannot be stained by the Gram stain procedure.
.mw-parser-output .tocleft{float:left;clear:left;width:auto;background:none;padding:.5em .8em 1.4em 0;margin-bottom:.5em}.mw-parser-output .tocleft-clear-left{clear:left}.mw-parser-output .tocleft-clear-both{clear:both}.mw-parser-output .tocleft-clear-none{clear:none}
Contents
1 Microbiologic characteristics
1.1 Metabolism and morphology
1.2 Pigmentation
1.3 Staining characteristics
2 Ecology
3 Pathogenicity
3.1 Medical classification
4 Mycosides
5 Genomics
6 Evolution
7 Species
7.1 Slowly growing
7.1.1 Mycobacterium tuberculosis complex
7.1.2 Mycobacterium avium complex
7.1.3 Mycobacterium gordonae clade
7.1.4 Mycobacterium kansasii clade
7.1.5 Mycobacterium nonchromogenicum/terrae clade
7.1.6 Mycolactone-producing mycobacteria
7.1.7 Mycobacterium simiae clade
7.1.8 Ungrouped
7.2 Intermediate growth rate
7.3 Rapidly growing
7.3.1 Mycobacterium abscessus clade
7.3.2 Mycobacterium chelonae clade
7.3.3 Mycobacterium fortuitum clade
7.3.4 Mycobacterium mucogenicum clade
7.3.5 Mycobacterium parafortuitum clade
7.3.6 Mycobacterium vaccae clade
7.3.7 CF
7.3.8 Ungrouped
7.4 Ungrouped
7.5 Proposed division of the genus
8 Mycobacteriophage
9 References
10 Further reading
11 External links
Microbiologic characteristics
Metabolism and morphology

Mycobacterial cell wall: 1-outer lipids, 2-mycolic acid, 3-polysaccharides (arabinogalactan), 4-peptidoglycan, 5-plasma membrane, 6-lipoarabinomannan (LAM), 7-phosphatidylinositol mannoside, 8-cell wall skeleton
Mycobacteria are aerobic. They are bacillary in form, at least in most phases that have attracted human microbiological attention to date; they are straight or slightly curved rods between 0.2 and 0.6 µm wide and between 1.0 and 10 µm long. They are generally nonmotile bacteria, except for the species Mycobacterium marinum, which has been shown to be motile within macrophages. They are characteristically acid-fast.[2] Mycobacteria have an outer membrane.[4] They possess capsules, and most do not form endospores. M. marinum and perhaps M. bovis have been shown to sporulate;[5] however, this has been contested by further research.[6] The distinguishing characteristic of all Mycobacterium species is that the cell wall is thicker than in many other bacteria, being hydrophobic, waxy, and rich in mycolic acids/mycolates. The cell wall consists of the hydrophobic mycolate layer and a peptidoglycan layer held together by a polysaccharide, arabinogalactan. The cell wall makes a substantial contribution to the hardiness of this genus. The biosynthetic pathways of cell wall components are potential targets for new drugs for tuberculosis.[7]
Many Mycobacterium species adapt readily to growth on very simple substrates, using ammonia or amino acids as nitrogen sources and glycerol as a carbon source in the presence of mineral salts. Optimum growth temperatures vary widely according to the species and range from 25 °C to over 50 °C.
Most Mycobacterium species, including most clinically relevant species, can be cultured in blood agar.[8] However, some species grow very slowly due to extremely long reproductive cycles — M. leprae, may take more than 20 days to proceed through one division cycle (for comparison, some E. coli strains take only 20 minutes), making laboratory culture a slow process.[2] In addition, the availability of genetic manipulation techniques still lags far behind that of other bacterial species.[9]
A natural division occurs between slowly– and rapidly–growing species. Mycobacteria that form colonies clearly visible to the naked eye within 7 days on subculture are termed rapid growers, while those requiring longer periods are termed slow growers.
Pigmentation
Some mycobacteria produce carotenoid pigments without light. Others require photoactivation for pigment production.
- Photochromogens (Group I)
- Produce nonpigmented colonies when grown in the dark and pigmented colonies only after exposure to light and reincubation.
- Ex: M. kansasii, M. marinum, M. simiae.
- Scotochromogens (Group II)
- Produce deep yellow to orange colonies when grown in the presence of either the light or the dark.
- Ex: M. scrofulaceum, M. gordonae, M. xenopi, M. szulgai.
- Non-chromogens (Groups III and IV)
- Nonpigmented in the light and dark or have only a pale yellow, buff or tan pigment that does not intensify after light exposure.
- Ex: M. tuberculosis, M. avium-intra-cellulare, M. bovis, M. ulcerans
- Ex: M. fortuitum, M. chelonae
- Ex: M. tuberculosis, M. avium-intra-cellulare, M. bovis, M. ulcerans
Staining characteristics
Mycobacteria are classical acid-fast organisms.[10] Stains used in evaluation of tissue specimens or microbiological specimens include Fite's stain, Ziehl-Neelsen stain, and Kinyoun stain.
Mycobacteria appear phenotypically most closely related to members of Nocardia, Rhodococcus, and Corynebacterium.
Ecology
Mycobacteria are widespread organisms, typically living in water (including tap water treated with chlorine) and food sources. Some, however, including the tuberculosis and the leprosy organisms, appear to be obligate parasites and are not found as free-living members of the genus.
Pathogenicity
Mycobacteria can colonize their hosts without the hosts showing any adverse signs. For example, billions of people around the world have asymptomatic infections of M. tuberculosis.
Mycobacterial infections are notoriously difficult to treat. The organisms are hardy due to their cell wall, which is neither truly Gram negative nor positive. In addition, they are naturally resistant to a number of antibiotics that disrupt cell-wall biosynthesis, such as penicillin. Due to their unique cell wall, they can survive long exposure to acids, alkalis, detergents, oxidative bursts, lysis by complement, and many antibiotics. Most mycobacteria are susceptible to the antibiotics clarithromycin and rifamycin, but antibiotic-resistant strains have emerged.
As with other bacterial pathogens, M. tuberculosis produces a number of surface and secreted proteins that contribute to its virulence. However, the mechanism by which these proteins contribute to virulence remains unknown.[11]
Medical classification
Mycobacteria can be classified into several major groups for purpose of diagnosis and treatment: M. tuberculosis complex, which can cause tuberculosis: M. tuberculosis, M. bovis, M. africanum, and M. microti; M. leprae, which causes Hansen's disease or leprosy; nontuberculous mycobacteria (NTM) are all the other mycobacteria, which can cause pulmonary disease resembling tuberculosis, lymphadenitis, skin disease, or disseminated disease.
Mycosides
Mycosides are phenolic alcohols (such as phenolphthiocerol) that were shown to be components of Mycobacterium glycolipids that are termed glycosides of phenolphthiocerol dimycocerosate.[12] Mycosides A and B have 18 and 20 carbon atoms, respectively.[13]
Genomics

Comparison of protein orthology in M. tuberculosis, M. leprae, and M. smegmatis, three major model systems in Mycobacterium research[14]
Comparative analyses of mycobacterial genomes have identified several conserved indels and signature proteins that are uniquely found in all sequenced species from the genus Mycobacterium.[15][16] Additionally, 14 proteins are found only in the species from the genera Mycobacterium and Nocardia, suggesting that these two genera are closely related.[16]
The genomes of some mycobacteria are quite large when compared to other bacteria. For instance, the genome of M. vulneris encodes 6,653 proteins, which is larger than that of small eukaryotes such as yeast (which encodes only ~6000 proteins).[17]
Evolution
M. ulcerans evolved from M. marinum.[18]
Species

Phylogenetic position of the tubercle bacilli within the genus Mycobacterium:
The blue triangle corresponds to tubercle bacilli sequences that are identical or differing by a single nucleotide. The sequences of the genus Mycobacterium that matched most closely to those of M. tuberculosis were retrieved from the BIBI database (http://pbil.univ-lyon.fr/bibi/) and aligned with those obtained for 17 smooth and MTBC strains. The unrooted neighbor-joining tree is based on 1,325 aligned nucleotide positions of the 16S rRNA gene. The scale gives the pairwise distances after Jukes-Cantor correction. Bootstrap support values higher than 90% are indicated at the nodes.
Phenotypic tests can be used to identify and distinguish different mycobacteria species and strains. In older systems, mycobacteria are grouped based upon their appearance and rate of growth. However, these are symplesiomorphies, and more recent classification is based upon cladistics. Over 100 species are currently recognised.
O'Neill and coworkers recently presented a comprehensive phylogenetic analysis based on an alignment of core genomes of 57 strains of bacteria, including all available mycobacteria.[19]
Slowly growing
Runyon's group I, II and III
Mycobacterium tuberculosis complex
Also see main article about Mycobacterium tuberculosis complex
Mycobacterium tuberculosis complex (MTBC) members are causative agents of human and animal tuberculosis. Species in this complex include:
- M. africanum
- M. bovis
- M. bovis BCG
- M. canetti
- M. caprae
- M. microti
- M. mungi
- M. orygis
- M. pinnipedii
- M. suricattae
M. tuberculosis, the major cause of human tuberculosis
Mycobacterium avium complex
Mycobacterium avium complex (MAC) is a group of species that, in a disseminated infection but not lung infection, used to be a significant cause of death in AIDS patients. The species M. indicus pranii appears to be basal in this complex.[20] Species in this complex include:
- M. avium
M. avium paratuberculosis, which has been implicated in Crohn's disease in humans and is the causative agent of Johne's disease in cattle and sheep
- M. avium silvaticum
- M. avium "hominissuis"
- M. colombiense
- M. indicus pranii
- M. intracellulare
Mycobacterium gordonae clade
- M. asiaticum
- M. gordonae
Mycobacterium kansasii clade
- M. gastri
- M. kansasii
Mycobacterium nonchromogenicum/terrae clade
- M. hiberniae
- M. icosiumassiliensis
- M. nonchromogenicum
- M. terrae
- M. triviale
Mycolactone-producing mycobacteria
M. ulcerans, which causes the "Buruli", or "Bairnsdale" ulcer
- M. pseudoshottsii
- M. shottsii
Mycobacterium simiae clade
- M. florentinum
- M. genavense
- M. heidelbergense
- M. interjectum
- M. kubicae
- M. lentiflavum
- M. montefiorense
- M. palustre
- M. parascrofulaceum
- M. simiae
- M. triplex
Ungrouped
- M. arabiense
- M. aromaticivorans
- M. aquaticum
- M. bacteremicum
- M. bohemicum
- M. botniense
- M. branderi
- M. celatum
- M. chimaera
- M. conspicuum
- M. cookii
- M. doricum
- M. farcinogenes
- M. haemophilum
- M. heckeshornense
- M. intracellulare
- M. lacus
M. leprae, which causes leprosy
- M. lepraemurium
M. lepromatosis, another (less significant) cause of leprosy, described in 2008- M. liflandii
- M. llatzerense
- M. malmoense
M. marinum, causes a rare disease called Aquarium granuloma.- M. neoaurum
- M. monacense
- M. murale
- M. nebraskense
- M. saskatchewanense
- M. sediminis
- M. scrofulaceum
- M. shimoidei
- M. szulgai
- Mycobacterium talmoniae
- M. tusciae
- M. xenopi
- M. yongonense
Intermediate growth rate
- M. intermedium
Rapidly growing
Mycobacterium abscessus clade
- M. abscessus
- M. bolletii
- M. massiliense
Together they are known as the M. abscessus complex
Mycobacterium chelonae clade
- M. chelonae
- M. immunogenum
- M. stephanolepidis
Mycobacterium fortuitum clade
- M. boenickei
- M. brisbanense
- M. cosmeticum
- M. fortuitum
- M. fortuitum subsp. acetamidolyticum
- M. houstonense
- M. mageritense
- M. neworleansense
- M. peregrinum
- M. porcinum
- M. senegalense
- M. septicum
Mycobacterium mucogenicum clade
- Mycobacterium aubagnese
- M. mucogenicum
- Mycobacterium phocaicum
Mycobacterium parafortuitum clade
- M. austroafricanum
- M. diernhoferi
- M. frederiksbergense
- M. hodleri
- M. neoaurum
- M. parafortuitum
Mycobacterium vaccae clade
- M. aurum
- M. vaccae
CF
- M. chitae
- M. fallax
Ungrouped
- M. agri
- M. aichiense
- M. alvei
- M. arupense
- M. barrassiae
- M. brumae
- M. canariasense
- M. chubuense
- M. conceptionense
- M. confluentis
- M. duvalii
- M. elephantis
- M. flavescens
- M. gadium
- M. gilvum
- M. hassiacum
- M. holsaticum
- M. iranicum
- M. komossense
- M. madagascariense
- M. massilipolynesiensis
- M. moriokaense
- M. obuense
- M. phlei
- M. psychrotolerans
- M. pulveris
- M. pyrenivorans
M. smegmatis
- M. goodii
- M. wolinskyi
- M. sphagni
- M. thermoresistibile
- M. vanbaalenii
Ungrouped
- M. arosiense
- M. aubagnense
- M. chlorophenolicum
- M. fluoroanthenivorans
- M. kumamotonense
- M. novocastrense
- M. parmense
- M. poriferae
- M. rhodesiae
- M. seoulense
- M. tokaiense
Proposed division of the genus
Gupta et al. have, based on the analysis of 150 species in the genus, proposed dividing Mycobacterium into five genera.[21] The proposed new genera are:
Mycobacterium based on the Tuberculosis-Simiae clade
Mycolicibacterium based on the Fortuitum-Vaccae clade
Mycolicibacter based on the Terrae clade
Mycolicibacillus based on the Triviale clade
Mycobacteroides based on the Abscessus-Chelonae clade
Wider acceptance of this proposal is awaited.
Mycobacteriophage
Mycobacteria can be infected by mycobacteriophages, bacterial viruses that may be used in the future to treat tuberculosis and related diseases by phage therapy.
The procedure may not go into practice in the case of Mtb for some time, as bacteriophage particles cannot penetrate into the tuberculosis bacilli, or clumps.
References
^ King HC, Khera-Butler T, James P, Oakley BB, Erenso G, Aseffa A, Knight R, Wellington EM, Courtenay O (2017) Environmental reservoirs of pathogenic mycobacteria across the Ethiopian biogeographical landscape. PLoS One 12(3):e0173811. doi: 10.1371/journal.pone.0173811
^ abc Ryan KJ, Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 0-8385-8529-9.CS1 maint: Extra text: authors list (link) .mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ James H. Kerr and Terry L. Barrett, "Atypical Mycobacterial Diseases", Military Dermatology Textbook, p. 401.
^ Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H (2010). "Mycobacterial outer membranes: in search of proteins". Trends in Microbiology. 18 (3): 109–16. doi:10.1016/j.tim.2009.12.005. PMC 2931330. PMID 20060722.
^ Ghosh, Jaydip; Larsson, Pontus; Singh, Bhupender; Pettersson, B M Fredrik; Islam, Nurul M; Nath Sarkar, Sailendra; Dasgupta, Santanu; Kirsebom, Leif A (2009). "Sporulation in mycobacteria". Proceedings of the National Academy of Sciences of the United States of America. 106 (26): 10781–10786. doi:10.1073/pnas.0904104106. PMC 2705590. PMID 19541637.
^ Traag, BA; Driks, A; Stragier, P; Bitter, W; Broussard, G; Hatfull, G; Chu, F; Adams, KN; Ramakrishnan, L; Losick, R (Jan 2010). "Do mycobacteria produce endospores?". Proc Natl Acad Sci U S A. 107 (2): 878–81. doi:10.1073/pnas.0911299107. PMC 2818926. PMID 20080769.
^ Bhamidi S (2009). "Mycobacterial Cell Wall Arabinogalactan". Bacterial Polysaccharides: Current Innovations and Future Trends. Caister Academic Press. ISBN 978-1-904455-45-5.
^ Lagier, Jean-Christophe; Edouard, Sophie; Pagnier, Isabelle; Mediannikov, Oleg; Drancourt, Michel; Raoult, Didier (2015). "Current and Past Strategies for Bacterial Culture in Clinical Microbiology". Clinical Microbiology Reviews. 28 (1): 208–36. doi:10.1128/CMR.00110-14. PMC 4284306. PMID 25567228.
^ Parish T, Brown A (editors) (2009). Mycobacterium: Genomics and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-40-0.CS1 maint: Extra text: authors list (link)
^ McMurray DN (1996). "Mycobacteria and Nocardia". In Baron S et al. (eds.). Baron's Medical Microbiology (4th ed.). Univ of Texas Medical Branch. ISBN 0-9631172-1-1.CS1 maint: Extra text: editors list (link)
^ McCann, Jessica R.; Kurtz, Sherry; Braunstein, Miriam (2009). "Secreted and Exported Proteins Important to Mycobacterium tuberculosis Pathogenesis". In Wooldridge, Karl. Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis. Norfolk, UK: Caister Academic Press. pp. 265–297. ISBN 9781904455424.
^ Smith, D.W., et al., Nature 1960, 186, 887
^ fatty alcohols and aldehydes
^ Akinola R. et al. 2013 A Systems Level Comparison of Mycobacterium tuberculosis, Mycobacterium leprae and Mycobacterium smegmatis Based on Functional Interaction. J Bacteriol Parasitol 2013, 4:4
^ Gao, B.; Paramanathan, R.; Gupta, R. S. (2006). "Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups". Antonie van Leeuwenhoek. 90 (1): 69–91. doi:10.1007/s10482-006-9061-2. PMID 16670965.
^ ab Gao, B.; Gupta, R. S. (2012). "Phylogenetic Framework and Molecular Signatures for the Main Clades of the Phylum Actinobacteria". Microbiology and Molecular Biology Reviews. 76 (1): 66–112. doi:10.1128/MMBR.05011-11. PMC 3294427. PMID 22390973.
^ Croce, Olivier; Robert, Catherine; Raoult, Didier; Drancourt, Michel (2014-05-08). "Draft Genome Sequence of Mycobacterium vulneris DSM 45247T". Genome Announcements. 2 (3). doi:10.1128/genomeA.00370-14. ISSN 2169-8287. PMC 4014686. PMID 24812218.
^ Vandelannoote K, Meehan CJ, Eddyani M, Affolabi D, Phanzu DM, Eyangoh S, Jordaens K, Portaels F, Mangas K, Seemann T, Marsollier L, Marion E, Chauty A, Landier J, Fontanet A, Leirs H, Stinear TP, de Jong BC1 (2017) Multiple Introductions and Recent Spread of the Emerging Human Pathogen Mycobacterium ulcerans across Africa. Genome Biol Evol 9(3):414-426
^ O'Neill, MB; Mortimer, TD; Pepperell, CS (2015). "Diversity of Mycobacterium tuberculosis across Evolutionary Scales". PLoS Pathog. 11 (11): e1005257. doi:10.1371/journal.ppat.1005257.
^ Rahman, SA; Singh, Y; Kohli, S; Ahmad, J; Ehtesham, NZ; Tyagi, AK; Hasnain, SE (2014). "Comparative analyses of nonpathogenic, opportunistic, and totally pathogenic Mycobacteria reveal genomic and biochemical variabilities and highlight the survival attributes of Mycobacterium tuberculosis". mBio. 5 (6): e02020–14. doi:10.1128/mBio.02020-14.
^ Gupta RS, Lo B, Son J (2018) Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Further reading
.mw-parser-output .refbegin{font-size:90%;margin-bottom:0.5em}.mw-parser-output .refbegin-hanging-indents>ul{list-style-type:none;margin-left:0}.mw-parser-output .refbegin-hanging-indents>ul>li,.mw-parser-output .refbegin-hanging-indents>dl>dd{margin-left:0;padding-left:3.2em;text-indent:-3.2em;list-style:none}.mw-parser-output .refbegin-100{font-size:100%}
Diagnosis and Treatment of Disease Caused by Nontuberculous Mycobacteria. American Thoracic Society. Am J Respiratory and Critical Care Medicine. Aug 1997 156(2) Part 2 Supplement
RIDOM: Ribosomal Differentiation of Medical Microorganisms- J.P. Euzéby: List of Prokaryotic Names with Standing in Nomenclature - Genus Mycobacterium
External links
Tuberculist: Genome annotation database
MTB Sysborg: Genome annotation database from the Institute of Genomics and Integrative Biology
TB Structural Genomics Consortium: Structures of Mycobacterium tuberculosis proteins
MycDB: Mycobacterium database
TBDB: Tuberculosis database
Mycobacterium genomes and related information at PATRIC, a Bioinformatics Resource Center funded by NIAID
- Frequently Asked Questions about NTM Lung Disease
- PRASITE: Identification of mycobacteria