Pilus
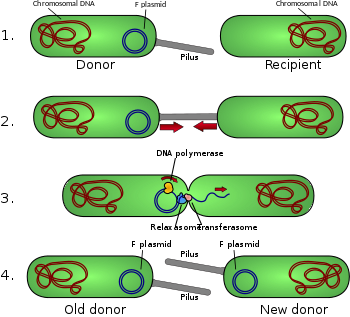
Schematic drawing of bacterial conjugation. 1- Donor cell produces pilus. 2- Pilus attaches to recipient cell, brings the two cells together. 3- The mobile plasmid is nicked and a single strand of DNA is then transferred to the recipient cell. 4- Both cells recircularize their plasmids, synthesize second strands, and reproduce pili; both cells are now viable donors.
A pilus (Latin for 'hair'; plural: pili) is a hair-like appendage found on the surface of many bacteria.[1] The terms pilus and fimbria (Latin for 'fringe'; plural: fimbriae) can be used interchangeably, although some researchers reserve the term pilus for the appendage required for bacterial conjugation. All pili in the latter sense are primarily composed of pilin proteins, which are oligomeric.
Dozens of these structures can exist on the bacteria. Some bacteria, viruses or bacteriophages attach to receptors on pili at the start of their reproductive cycle.
Pili are antigenic. They are also fragile and constantly replaced, sometimes with pili of different composition, resulting in altered antigenicity. Specific host responses to old pili structure are not effective on the new structure. Recombination genes of pili code for variable (V) and constant (C) regions of the pili (similar to immunoglobulin diversity).
Contents
1 Types
1.1 Conjugative pili
1.2 Type IV pili
2 Fimbriae
3 Virulence
4 See also
5 References
6 External links
Types
Conjugative pili
Conjugative pili allow for the transfer of DNA between bacteria, in the process of bacterial conjugation. They are sometimes called "sex pili", in analogy to sexual reproduction, because they allow for the exchange of genes via the formation of "mating pairs". Perhaps the most well-studied is the pilus of Escherichia coli, encoded by the fertility F sex factor.
A pilus is typically 6 to 7 nm in diameter. During conjugation, a pilus emerging from the donor bacterium ensnares the recipient bacterium, draws it in close, and eventually triggers the formation of a mating bridge, which establishes direct contact and the formation of a controlled pore that allows transfer of DNA from the donor to the recipient. Typically, the DNA transferred consists of the genes required to make and transfer pili (often encoded on a plasmid), and so is a kind of selfish DNA; however, other pieces of DNA are often co-transferred and this can result in dissemination of genetic traits throughout a bacterial population, such as antibiotic resistance. Not all bacteria can make conjugative pili, but conjugation can occur between bacteria of different species.[citation needed]
Type IV pili

Type IV Pilus Twitching Motility 1. Pre-PilA is made in the cytoplasm and moves into the inner membrane. 2. Pre-PilA is inserted into the inner membrane. 3. PilD, a peptidase, removes a leader sequence, thus making the Pre-PilA shorter and into PilA, the main building-block protein of Pili. 4. PilF, a NTP-Binding protein that provides energy for Type IV Pili Assembly. 5. The secretin protein, PilQ, found on the outer membrane of the cell is necessary for the development/extension of the pilus. PilC is the first proteins to form the pilus and are responsible for overall attachment of the pilus. 6. Once the Type IV Pilus attaches or interacts with what it needs to, it begins to retract. This occurs with the PilT beginning to degrade the last parts of the PilA in the pilus. The mechanism of PilT is very similar to PilF. 7. Degradation of the pilus into the components to be utilized and synthesized into PilA again.[2]
Some pili, called type IV pili (T4P), generate motile forces.[3] The external ends of the pili adhere to a solid substrate, either the surface to which the bacterium is attached or to other bacteria. Then, when the pili contract, they pull the bacterium forward like a grappling hook. Movement produced by type IV pili is typically jerky, so it is called twitching motility, as opposed to other forms of bacterial motility such as that produced by flagella. However, some bacteria, for example Myxococcus xanthus, exhibit gliding motility. Bacterial type IV pili are similar in structure to the component flagellins of archaella (archaeal flagella).[4]
Genetic transformation is the process by which a recipient bacterial cell takes up DNA from a neighboring cell and integrates this DNA into its genome by homologous recombination. In Neisseria meningitidis (also called meningococcus), DNA transformation requires the presence of short DNA uptake sequences (DUSs) which are 9-10 monomers residing in coding regions of the donor DNA. Specific recognition of DUSs is mediated by a type IV pilin.[5] Menningococcal type IV pili bind DNA through the minor pilin ComP via an electropositive stripe that is predicted to be exposed on the filament's surface. ComP displays an exquisite binding preference for selective DUSs. The distribution of DUSs within the N. meningitides genome favors certain genes, suggesting that there is a bias for genes involved in genomic maintenance and repair.[6][7]
Fimbriae
To initiate formation of a biofilm, fimbriae must attach bacteria to host surfaces for colonization during infection. A fimbria is a short pilus that is used to attach the bacterium to a surface. They are sometimes called "attachment pili". Fimbriae are either located at the poles of a cell or are evenly spread over its entire surface. Mutant bacteria that lack fimbriae cannot adhere to their usual target surfaces, and thus cannot cause diseases.
Some fimbriae can contain lectins. The lectins are necessary to adhere to target cells because they can recognize oligosaccharide units on the surface of these target cells. Other fimbriae bind to components of the extracellular matrix. Fimbriae found in Gram-negative have the pilin subunits covalently linked.
Some aerobic bacteria form a thin layer at the surface of a broth culture. This layer, called a pellicle, consists of many aerobic bacteria that adhere to the surface by their fimbriae or "attachment pili". Thus, fimbriae allow the aerobic bacteria to remain on the broth, from which they take nutrients, while they congregate near the air.[citation needed]
Virulence
Pili are responsible for virulence in the pathogenic strains of many bacteria, including E. coli, Vibrio cholerae, and many strains of Streptococcus.[8][9] This is because the presence of pili greatly enhances bacteria's ability to bind to body tissues, which then increases replication rates and ability to interact with the host organism.[8] If a species of bacteria has multiple strains but only some are pathogenic, it is likely that the pathogenic strains will have pili while the nonpathogenic strains won't.[10][11]
The development of attachment pili may then result in the development of further virulence traits. Nonpathogenic strains of V. cholerae first evolved pili, allowing them to bind to human tissues and form microcolonies.[8][11] These pili then served as binding sites for the lysogenic bacteriophage that carries the disease-causing toxin.[8][11] The gene for this toxin, once incorporated into the bacterium's genome, is expressed when the gene coding for the pilus is expressed (hence the name "toxin mediated pilus").[8]
See also
- Bacterial nanowires
- Flagellum
- Sortase
References
^ "pilus" at Dorland's Medical Dictionary
^ Joan, Slonczewski (2017). Microbiology : an evolving science. Foster, John Watkins (Fourth ed.). New York: W. W. Norton & Company. pp. 1000–1002. ISBN 9780393614039. OCLC 951925510..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output .citation q{quotes:"""""""'""'"}.mw-parser-output .citation .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-limited a,.mw-parser-output .citation .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-ws-icon a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/Wikisource-logo.svg/12px-Wikisource-logo.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-maint{display:none;color:#33aa33;margin-left:0.3em}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Mattick JS (2002). "Type IV pili and twitching motility". Annu. Rev. Microbiol. 56 (1): 289–314. doi:10.1146/annurev.micro.56.012302.160938. PMID 12142488.
^ Jarrell; et al. (2009). "Archaeal Flagella and Pili". Pili and Flagella: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-48-6.
^ Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V (2013). "Specific DNA recognition mediated by a type IV pilin". Proc. Natl. Acad. Sci. U.S.A. 110 (8): 3065–70. doi:10.1073/pnas.1218832110. PMC 3581936. PMID 23386723.
^ Davidsen T, Rødland EA, Lagesen K, Seeberg E, Rognes T, Tønjum T (2004). "Biased distribution of DNA uptake sequences towards genome maintenance genes". Nucleic Acids Res. 32 (3): 1050–8. doi:10.1093/nar/gkh255. PMC 373393. PMID 14960717.
^ Caugant DA, Maiden MC (2009). "Meningococcal carriage and disease--population biology and evolution". Vaccine. 27 Suppl 2: B64–70. doi:10.1016/j.vaccine.2009.04.061. PMC 2719693. PMID 19464092.
^ abcde Craig, Lisa; Taylor, Ronald (2014). "Chapter 1: The Vibrio cholerae Toxin Coregulated Pilus: Structure, Assembly, and Function with Implications for Vaccine Design". In Barocchi, Michèle; Telford, John. Bacterial Pili: Structure, Synthesis, and Role in Disease. C.A.B. International. pp. 1–16. ISBN 978-1-78064-255-0.
^ Rinaudo, Daniela; Moschioni, Monica (2014). "Chapter 13: Pilus-based Vaccine Development in Streptococci: Variability, Diversity, and Immunological Resposes". In Barocchi, Michèle; Telford, John. Bacterial Pili: Structure, Synthesis, and Role in Disease. C.A.B. International. pp. 182–202. ISBN 978-1-78064-255-0.
^ Todar, Kenneth. "Textbook of Bacteriology: Bacterial Structure in Relationship to Pathogenicity". Textbook of Bacteriology. Retrieved 24 November 2017.
^ abc Georgiadou, Michaella; Pelicic, Vladimir (2014). "Chapter 5: Type IV Pili: Functions & Biogenesis". In Barocchi, Michèle; Telford, John. Bacterial Pili: Structure, Synthesis, and Role in Disease. C.A.B. International. pp. 71–84. ISBN 978-1-78064-255-0.
External links
Sex+Pilus at the US National Library of Medicine Medical Subject Headings (MeSH)
Bacterial+Pilus at the US National Library of Medicine Medical Subject Headings (MeSH)