Mycobacterium tuberculosis sRNA

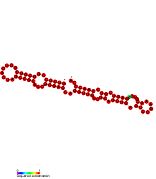
Secondary structure of b55, one of the sRNAs experimentally confirmed in M. tuberculosis
Mycobacterium tuberculosis contains at least nine small RNA families in its genome.[1] The small RNA (sRNA) families were identified through RNomics – the direct analysis of RNA molecules isolated from cultures of Mycobacterium tuberculosis.[2][3] The sRNAs were characterised through RACE mapping and Northern blot experiments.[1]Secondary structures of the sRNAs were predicted using Mfold.[4]
sRNAPredict2 – a bioinformatics tool – suggested 56 putative sRNAs in M. tuberculosis, though these have yet to be verified experimentally.[5]Hfq protein homologues have yet to be found in M. tuberculosis;[6] an alternative pathway – potentially involving conserved C-rich motifs – has been theorised to enable trans-acting sRNA functionality.[1]
sRNAs were shown to have important physiological roles in M. tuberculosis. Overexpression of G2 sRNA, for example, prevented growth of M. tuberculosis and greatly reduced the growth of M. smegmatis; ASdes sRNA is thought to be a cis-acting regulator of a fatty acid desaturase (desA2) while ASpks is found with the open reading frame for Polyketide synthase-12 (pks12) and is an antisense regulator of pks12 mRNA.[1]
The sRNA ncrMT1302 was found to be flanked by the MT1302 and MT1303 open reading frames. MT1302 encodes an adenylyl cyclase that converts ATP to cAMP, the expression of ncrMT1302 is regulated by cAMP and pH.[7]
Mcr7 sRNA encoded by the mcr7 gene modulates translation of the tatC mRNA and impacts the activity of the Twin Arginine Translocation (Tat) protein secretion apparatus.[8]
npcTB_6715 is a first sRNA identified as a potential biomarker for the detection of MTB in patients.[9]
See also
Bacillus subtilis sRNAs- Bacterial small RNA
Brucella sRNA
Caenorhabditis elegans sRNA
Escherichia coli sRNA
Pseudomonaa sRNA
References
^ abcd Arnvig KB, Young DB (August 2009). "Identification of small RNAs in Mycobacterium tuberculosis". Molecular Microbiology. 73 (3): 397–408. doi:10.1111/j.1365-2958.2009.06777.x. PMC 2764107. PMID 19555452..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, Hüttenhofer A, Wagner EG (November 2003). "RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria". Nucleic Acids Research. 31 (22): 6435–43. doi:10.1093/nar/gkg867. PMC 275561. PMID 14602901.
^ Kawano M, Reynolds AA, Miranda-Rios J, Storz G (2005). "Detection of 5'- and 3'-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli". Nucleic Acids Research. 33 (3): 1040–50. doi:10.1093/nar/gki256. PMC 549416. PMID 15718303.
^ Zuker M (July 2003). "Mfold web server for nucleic acid folding and hybridization prediction". Nucleic Acids Research. 31 (13): 3406–15. doi:10.1093/nar/gkg595. PMC 169194. PMID 12824337.
^ Livny J, Brencic A, Lory S, Waldor MK (2006). "Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2". Nucleic Acids Research. 34 (12): 3484–93. doi:10.1093/nar/gkl453. PMC 1524904. PMID 16870723.
[dead link]
^ Sun X, Zhulin I, Wartell RM (September 2002). "Predicted structure and phyletic distribution of the RNA-binding protein Hfq". Nucleic Acids Research. 30 (17): 3662–71. doi:10.1093/nar/gkf508. PMC 137430. PMID 12202750.
^ Pelly S, Bishai WR, Lamichhane G (May 2012). "A screen for non-coding RNA in Mycobacterium tuberculosis reveals a cAMP-responsive RNA that is expressed during infection". Gene. 500 (1): 85–92. doi:10.1016/j.gene.2012.03.044. PMC 3340464. PMID 22446041.
^ Solans L, Gonzalo-Asensio J, Sala C, Benjak A, Uplekar S, Rougemont J, Guilhot C, Malaga W, Martín C, Cole ST (May 2014). "The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis". PLoS Pathogens. 10 (5): e1004183. doi:10.1371/journal.ppat.1004183. PMC 4038636. PMID 24874799.
^ Kanniappan P, Ahmed SA, Rajasekaram G, Marimuthu C, Ch'ng ES, Lee LP, Raabe CA, Rozhdestvensky TS, Tang TH (October 2017). "RNomic identification and evaluation of npcTB_6715, a non-protein-coding RNA gene as a potential biomarker for the detection of Mycobacterium tuberculosis". Journal of Cellular and Molecular Medicine. 21 (10): 2276–2283. doi:10.1111/jcmm.13148. PMC 5618688. PMID 28756649.
Further reading
.mw-parser-output .refbegin{font-size:90%;margin-bottom:0.5em}.mw-parser-output .refbegin-hanging-indents>ul{list-style-type:none;margin-left:0}.mw-parser-output .refbegin-hanging-indents>ul>li,.mw-parser-output .refbegin-hanging-indents>dl>dd{margin-left:0;padding-left:3.2em;text-indent:-3.2em;list-style:none}.mw-parser-output .refbegin-100{font-size:100%}
Pánek J, Bobek J, Mikulík K, Basler M, Vohradský J (May 2008). "Biocomputational prediction of small non-coding RNAs in Streptomyces". BMC Genomics. 9: 217. doi:10.1186/1471-2164-9-217. PMC 2422843. PMID 18477385.
Livny J, Waldor MK (April 2007). "Identification of small RNAs in diverse bacterial species". Current Opinion in Microbiology. 10 (2): 96–101. doi:10.1016/j.mib.2007.03.005. PMID 17383222.
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. (June 1998). "Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence". Nature. 393 (6685): 537–44. doi:10.1038/31159. PMID 9634230.
Arnvig KB, Gopal B, Papavinasasundaram KG, Cox RA, Colston MJ (February 2005). "The mechanism of upstream activation in the rrnB operon of Mycobacterium smegmatis is different from the Escherichia coli paradigm". Microbiology. 151 (Pt 2): 467–73. doi:10.1099/mic.0.27597-0. PMID 15699196.
Matsunaga I, Bhatt A, Young DC, Cheng TY, Eyles SJ, Besra GS, Briken V, Porcelli SA, Costello CE, Jacobs WR, Moody DB (December 2004). "Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells". The Journal of Experimental Medicine. 200 (12): 1559–69. doi:10.1084/jem.20041429. PMC 2211992. PMID 15611286.