Variable number tandem repeat
A variable number tandem repeat (or VNTR) is a location in a genome where a short nucleotide sequence is organized as a tandem repeat. These can be found on many chromosomes, and often show variations in length (number of repeats) among individuals. Each variant acts as an inherited allele, allowing them to be used for personal or parental identification. Their analysis is useful in genetics and biology research, forensics, and DNA fingerprinting.
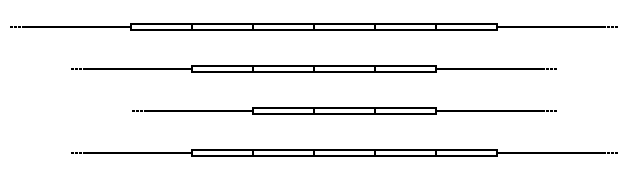
Schematic of a Variable Number of Tandem Repeats in 4 alleles.
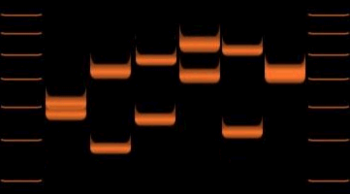
Variations of VNTR (D1S80) allele lengths in 6 individuals.
Contents
1 Structure and allelic variation
2 Use in genetic analysis
3 Inheritance
4 Relationship to other types of repetitive DNA
5 Classes
6 See also
6.1 Software for MLVA typing
7 References
8 External links
Structure and allelic variation
In the schematic above, the rectangular blocks represent each of the repeated DNA sequences at a particular VNTR location. The repeats are in tandem – i.e. they are clustered together and oriented in the same direction. Individual repeats can be removed from (or added to) the VNTR via recombination or replication errors, leading to alleles with different numbers of repeats. Flanking the repeats are segments of non-repetitive sequence (shown here as thin lines), allowing the VNTR blocks to be extracted with restriction enzymes and analyzed by RFLP, or amplified by the polymerase chain reaction (PCR) technique and their size determined by gel electrophoresis.
Use in genetic analysis
VNTRs were an important source of RFLP genetic markers used in linkage analysis (mapping) of diploid genomes. Now that many genomes have been sequenced, VNTRs have become essential to forensic crime investigations, via DNA fingerprinting and the CODIS database. When removed from surrounding DNA by the PCR or RFLP methods, and their size determined by gel electrophoresis or Southern blotting, they produce a pattern of bands unique to each individual. When tested with a group of independent VNTR markers, the likelihood of two unrelated individuals' having the same allelic pattern is extremely low. VNTR analysis is also being used to study genetic diversity and breeding patterns in populations of wild or domesticated animals. As such, VNTRs can be used to distinguish strains of bacterial pathogens. In this microbial forensics context, such assays are usually called Multiple Loci VNTR Analysis or MLVA.

Chromosomal locations of the 13 VNTR loci in the CODIS panel.
Inheritance
In analyzing VNTR data, two basic genetic principles can be used:
Identity Matching – both VNTR alleles from a specific location must match. If two samples are from the same individual, they must show the same allele pattern.
Inheritance Matching – the VNTR alleles must follow the rules of inheritance. In matching an individual with his parents or children, a person must have an allele that matches one from each parent. If the relationship is more distant, such as a grandparent or sibling, then matches must be consistent with the degree of relatedness.
Relationship to other types of repetitive DNA
Repetitive DNA, representing over 40% of the human genome, is arranged in a bewildering array of patterns. Repeats were first identified by the extraction of Satellite DNA, which does not reveal how they are organized. The use of restriction enzymes showed that some repeat blocks were interspersed throughout the genome. DNA sequencing later showed that other repeats are clustered at specific locations, with tandem repeats being more common than inverted repeats (which may interfere with DNA replication). VNTRs are the class of clustered tandem repeats that exhibit allelic variation in their lengths.
Classes

This shows a theoretical example of a VNTR in two different individuals. A single strand of DNA from each individual is displayed in which there is tandem repeat sequence that the individuals share. The sequence presence is a VNTR because one individual has five repeats, while the other has seven repeats (number of repeats varies in different individuals). Each repeat is ten nucleotides, making it a minisatellite, rather than a microsatellite in which each repeat is 2-5 nucleotides.
VNTRs are a type of minisatellite in which the size of the repeat sequence is generally ten to one hundred base pairs. Minisatellites are a type of DNA tandem repeat sequence, meaning that the sequences repeat one after another without other sequences or nucleotides in between them. Minisatellites are characterized by a repeat sequence of about ten to one hundred nucleotides, and the number of times the sequence repeats varies from about five to fifty times. The sequences of minisatellites are larger than those of microsatellites, in which the repeat sequence is generally two to five nucleotides. The two types of repeat sequences are both tandem but are specified by the length of the repeat sequence. VNTRs, therefore, because they have repeat sequences of ten to one hundred nucleotides in which every repeat is exactly the same, are considered minisatellites. However, while all VNTRs are minisatellites, not all minisatellites are VNTRs. VNTRs can vary in number of repeats from individual to individual, as where some non-VNTR minisatellites have repeat sequences that repeat the same number of times in all individuals containing the tandem repeats in the their genetic codes.[1][2]
See also
- AFLP
- Microsatellite
- Minisatellite
- MLVA
- Short tandem repeat
- Tandem repeat
Software for MLVA typing
BioNumerics (using the BioNumerics MIRU-VNTR plugin specifically developed for running a popular MLVA assay genotyping Mycobacterium tuberculosis strains)
References
^ https://www.nlm.nih.gov/visibleproofs/education/dna/vntr.pdf
^ https://www2.le.ac.uk/departments/mathematics/redundant-content/event-microsites/bio/Dubrova.pdf
External links
- Examples :
VNTRs – info and animated example
- Databases :
- The Microorganisms Tandem Repeats Database
- The MLVAbank
- Short Tandem Repeats Database
- Tandem Repeats Database (TRDB)
- Search tools :
- TAPO: A combined method for the identification of tandem repeats in protein structures
- Tandem Repeats Finder
- Mreps
- STAR
- TRED
- TandemSWAN
- Microsatellite repeats finder
- JSTRING – Java Search for Tandem Repeats in genomes
- Phobos – a tandem repeat search tool for perfect and imperfect repeats – the maximum pattern size depends only on computational power
Variable+Number+of+Tandem+Repeats at the US National Library of Medicine Medical Subject Headings (MeSH)